Code
library(gt)
library(tidyverse)
library(patchwork)
library(bggjphd)
theme_set(theme_bggj())A simulation study of the differences between AR(1) copula and i.i.d copula
Another step in my PhD studies is to apply multivariate copulas to data with Generalized Extreme Value marginal distributions. This is important to enable us to model dependence on the data leve, as opposed to the latent (parameter) level.
February 6, 2024
In a previous post, I wrote about applying the BYM2 model to spatially dependent Generalized Extreme Value (GEV) data and mentioned in passing that I was working on applying copulas as well. In this post I will outline and show results from a simulation study I’ve been running where I:
In the book Elements of Copula Modeling with R, Hofert et al. (2018) define a copula as
a multivariate distribution function with standard uniform univariate margins, that is, U(0, 1) margins.
So, a Copula is any multivariate distribution whose margins is U(0,1 ) distributed. A simple example is the independence copula
\[ \Pi(u) = \prod_{j=1}^{d}{u_j}, \quad \mathbf U \in [0, 1]^d. \]
The central theorem of copula theory is Sklar’s theorem (Sklar 1996) [the original paper is from 1959]. The theorem basically states that for any d-dimensional distribution function, \(H\), with univariate margins \(F_1, \dots, F_d\), there exists a d-dimensional copula \(C\) such that
\[ H(\mathbf x) = C(F_1(x_1), \dots, F_d(x_d)). \]
Alternatively, we can write
\[ C(\mathbf u) = H(F_1^{-1}(u_1), \dots, F_d^{-1}(u_d)) \]
The Gaussian family of copulas can be written
\[ C(\mathbf u | R) = \Phi_d\left(\Phi^{-1}(u_1), \dots, \Phi^{-1}(u_d) \vert R \right), \quad \mathbf u \in [0, 1]^d, \]
where R is a \(d\times d\) correlation matrix, \(\Phi_d\) is the CDF of a d-dimensional multivariate Gaussian with mean \(\mathbf 0\) and covariance matrix \(R\), and \(\Phi\) is the CDF of a standard Gaussian.
Its density is
\[ c(\mathbf u \vert R) = \frac{\phi_d\left(\Phi^{-1}(u_1), \dots, \Phi^{-1}(u_d) \vert R \right)}{ \prod_{j=1}^{d}{\phi\left( \Phi^{-1}(u_j) \right)}}, \quad \mathbf u \in [0, 1]^d, \]
where \(\phi_d\) is the density of the same multivariate Gaussian, and \(\phi\) is the density of a standard gaussian.
Given the \(d\times d\) correlation matrix, \(R\):
We can then make the margins \(u_1, \dots, u_d\) follow any distribution (f.ex. the GEV distribution) by applying its quantile function.
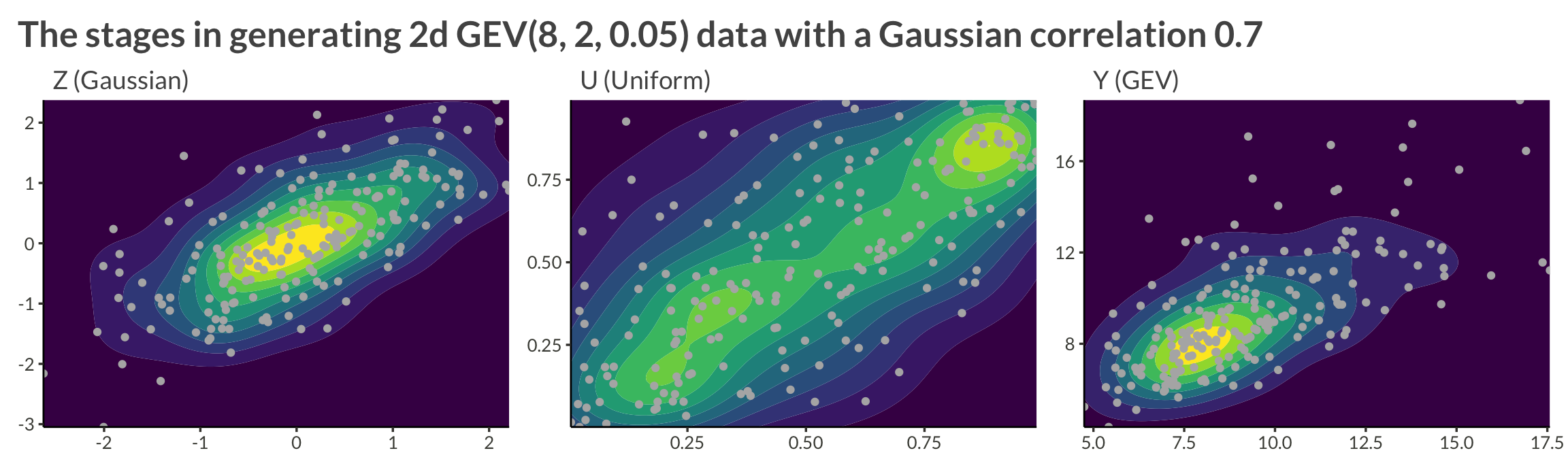
Let’s sample from a two-dimensional process with GEV(8, 2, 0.05) margins and a Gaussian copula with correlation \(\rho = 0.7\).
R <- matrix(c(1, 0.7, 0.7, 1), nrow = 2)
set.seed(1)
X <- mvtnorm::rmvnorm(
n = 200,
sigma = R
)
X |>
as_tibble() |>
mutate(
id = row_number()
) |>
pivot_longer(c(-id), names_to = "variable", values_to = "Z") |>
mutate(
U = pnorm(Z),
Y = evd::qgev(U, loc = 8, scale = 2, shape = 0.05)
) |>
pivot_longer(c(-id, -variable)) |>
pivot_wider(names_from = variable, values_from = value) |>
mutate(
name = fct_relevel(name, "Z", "U", "Y") |>
fct_recode(
"Z (Gaussian)" = "Z",
"U (Uniform)" = "U",
"Y (GEV)" = "Y"
)
) |>
group_by(name2 = name) |>
group_map(
function(data, ...) {
data |>
ggplot(aes(V1, V2)) +
geom_density_2d_filled() +
geom_point(col = "grey70") +
coord_cartesian(expand = FALSE) +
theme(legend.position = "none") +
labs(
x = NULL,
y = NULL,
subtitle = data$name
)
}
) |>
wrap_plots() +
plot_annotation(
title = "The stages in generating 2d GEV(8, 2, 0.05) data with a Gaussian correlation 0.7"
)
R scripts used for simulating the data
The simulated data have GEV margins and an AR(1) Gaussian copula. The steps in simulating the data are as follows.
First, a correlation matrix is generated that encodes an AR(1) process with correlation \(\rho\). To create this correlation matrix, we specify the precision matrix, \(Q\), via
\[ Q = \frac{1}{1 - \rho^2}\begin{bmatrix} 1 & -\rho & \dots & \dots & \vdots\\ -\rho & 1 + \rho^2 & -\rho & \ddots & \vdots \\ \vdots & \ddots & \ddots & \ddots & \vdots \\ \vdots & \ddots &-\rho & 1 + \rho^2 & -\rho \\ \vdots & \dots & \dots & -\rho & 1 \end{bmatrix}. \]
This gives us the correlation matrix, R, where
\[ R = Q^{-1} = \begin{bmatrix} 1 & \rho & \rho^2 & \dots & \rho^n\\ \rho & 1 & \rho & \ddots & \rho^{n-1} \\ \vdots & \ddots & \ddots & \ddots & \vdots \\ \rho^{n-1} & \ddots &\rho & 1 & \rho \\ \rho^n & \dots & \dots & \rho & 1 \end{bmatrix}. \]
make_AR_cor_matrix_1d <- function(n_id, rho = 0.5) {
P <- matrix(
0,
nrow = n_id,
ncol = n_id
)
diag(P) <- 1
for (i in seq(1, n_id - 1)) {
P[i, i + 1] <- -rho
}
for (i in seq(2, n_id)) {
P[i, i - 1] <- -rho
}
for (i in seq(2, n_id - 1)) {
P[i, i] <- 1 + rho^2
}
P <- P / (1 - rho^2)
P_cor <- solve(P)
P_cor
}
n_id <- 5
rho <- 0.5
R <- make_AR_cor_matrix_1d(n_id = n_id, rho = rho)
R [,1] [,2] [,3] [,4] [,5]
[1,] 1.0000 0.500 0.25 0.125 0.0625
[2,] 0.5000 1.000 0.50 0.250 0.1250
[3,] 0.2500 0.500 1.00 0.500 0.2500
[4,] 0.1250 0.250 0.50 1.000 0.5000
[5,] 0.0625 0.125 0.25 0.500 1.0000We then use the well-known fact that if
\[ \mathbf X \sim \mathrm{MVNorm}\left(\boldsymbol \mu, \Sigma\right), \]
then
\[ \boldsymbol X = \boldsymbol \mu + L \boldsymbol Z, \]
where \(\boldsymbol Z \sim \mathrm{Normal}(\boldsymbol 0, I)\) and \(L\) is the Cholesky decomposition of \(R\), or \(LL^T = \Sigma\).
In our case, we want \(\mathbf X\) to have mean 0 and covariance matrix \(R\), so we write
\[ \mathbf X = L\mathbf Z \]
where \(LL^T = R\).
We can skip the manual Cholesky factorization and simply use the {mvtnorm} package.
[,1] [,2] [,3] [,4] [,5]
[1,] 1.427875 1.837133 -0.1348893 -0.3970323 -0.4613493We will want more than one observation from each “site”, so we write a helped function for tidying this output into a tibble.
tidy_mvgauss <- function(mvnorm_matrix) {
colnames(mvnorm_matrix) <- seq_len(ncol(mvnorm_matrix))
mvnorm_matrix |>
dplyr::as_tibble() |>
dplyr::mutate(
replicate = dplyr::row_number(),
.before = `1`
) |>
tidyr::pivot_longer(
c(-replicate),
names_to = "id", names_transform = as.numeric,
values_to = "Z"
)
}
n_replicates <- 10
mvnorm_data <- R |>
sample_gaussian_variables(n_replicates = n_replicates) |>
tidy_mvgauss()
mvnorm_data |>
filter(replicate <= 5) |>
pivot_wider(names_from = replicate, values_from = Z) |>
gt() |>
tab_spanner(
columns = -id,
label = "Replicate"
)| id | Replicate | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| 1 | -0.4364007 | -0.1443106 | 0.7080631 | 1.1388723 | 1.0330390 |
| 2 | -0.4017539 | 0.6923541 | 0.9544816 | -0.4595604 | -1.3986486 |
| 3 | -0.1161437 | 0.9328314 | -0.1228664 | -0.4689164 | -1.1884398 |
| 4 | 1.0500406 | -0.5297770 | -0.8530328 | -1.5227378 | -1.0434809 |
| 5 | -0.3607882 | -0.5568922 | 1.3144552 | 0.1006016 | -0.9255078 |
Now our data, \(\mathbf X\), is multivariate normal with dependence structure according to an AR(1) process, but marginally each \(X_1\) is standard normal. To transform this data to a GEV dataset, we simply use the standard normal CDF on each \(X_i\) to transform it to \([0, 1]\), then we use the GEV quantile function to transform that to a GEV distributed variable.
mvnorm_to_gev <- function(mvnorm_data, gev_params) {
mvnorm_data |>
dplyr::mutate(
U = pnorm(Z)
) |>
dplyr::inner_join(
gev_params,
by = dplyr::join_by(id)
) |>
dplyr::mutate(
y = purrr:::pmap_dbl(
list(U, mu, sigma, xi),
\(U, mu, sigma, xi) evd::qgev(p = U, loc = mu, scale = sigma, shape = xi)
)
)
}
gev_params <- expand_grid(
mu = 6,
sigma = 3,
xi = 0.1,
id = seq_len(n_id)
)
gev_data <- mvnorm_data |>
mvnorm_to_gev(gev_params = gev_params)
gev_data |>
select(-mu, -sigma, -xi) |>
gt() |>
opt_interactive()Let’s create a function that plots heat maps for each of the three representations of the data
We want the color scales to be free, so we use group_map() and wrap_plots() instead of facet_wrap()
plot_data <- function(data) {
data |>
select(replicate, id, Z, U, y) |>
pivot_longer(c(-replicate, -id)) |>
mutate(
name = fct_relevel(name, "Z", "U", "y"),
name2 = name
) |>
group_by(name) |>
group_map(
function(data, ...) {
data |>
ggplot(aes(id, replicate, fill = value)) +
geom_raster() +
scale_fill_viridis_c() +
coord_cartesian(expand = FALSE) +
theme(legend.position = "none") +
labs(
subtitle = data$name2
)
}
) |>
wrap_plots() +
plot_layout(nrow = 1) +
plot_annotation(
title = "Heatmaps of the three data representations",
subtitle = "Z: Normal | U: Uniform | y: GEV"
)
}It’s nice to wrap this process in a single function
make_data <- function(gev_params, n_replicate, rho) {
make_AR_cor_matrix_1d(n_id = nrow(gev_params), rho = rho) |>
sample_gaussian_variables(n_replicates = n_replicate) |>
tidy_mvgauss() |>
dplyr::mutate(
U = pnorm(Z)
) |>
dplyr::inner_join(
gev_params,
by = dplyr::join_by(id)
) |>
dplyr::mutate(
y = purrr::pmap_dbl(
list(U, mu, sigma, xi),
\(U, mu, sigma, xi) evd::qgev(p = U, loc = mu, scale = sigma, shape = xi)
)
)
}Let’s see how the data look if we make it larger and increase the correlation
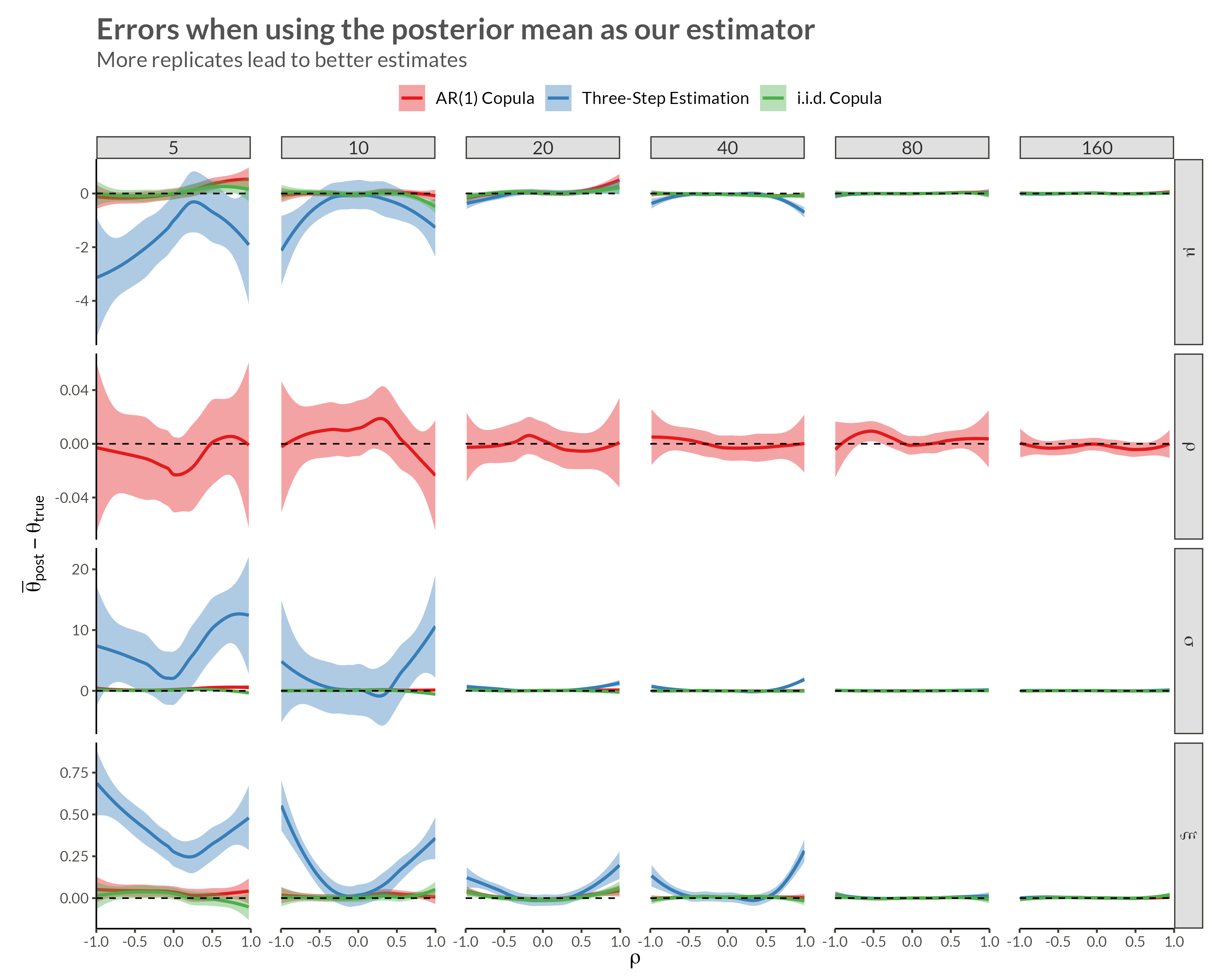
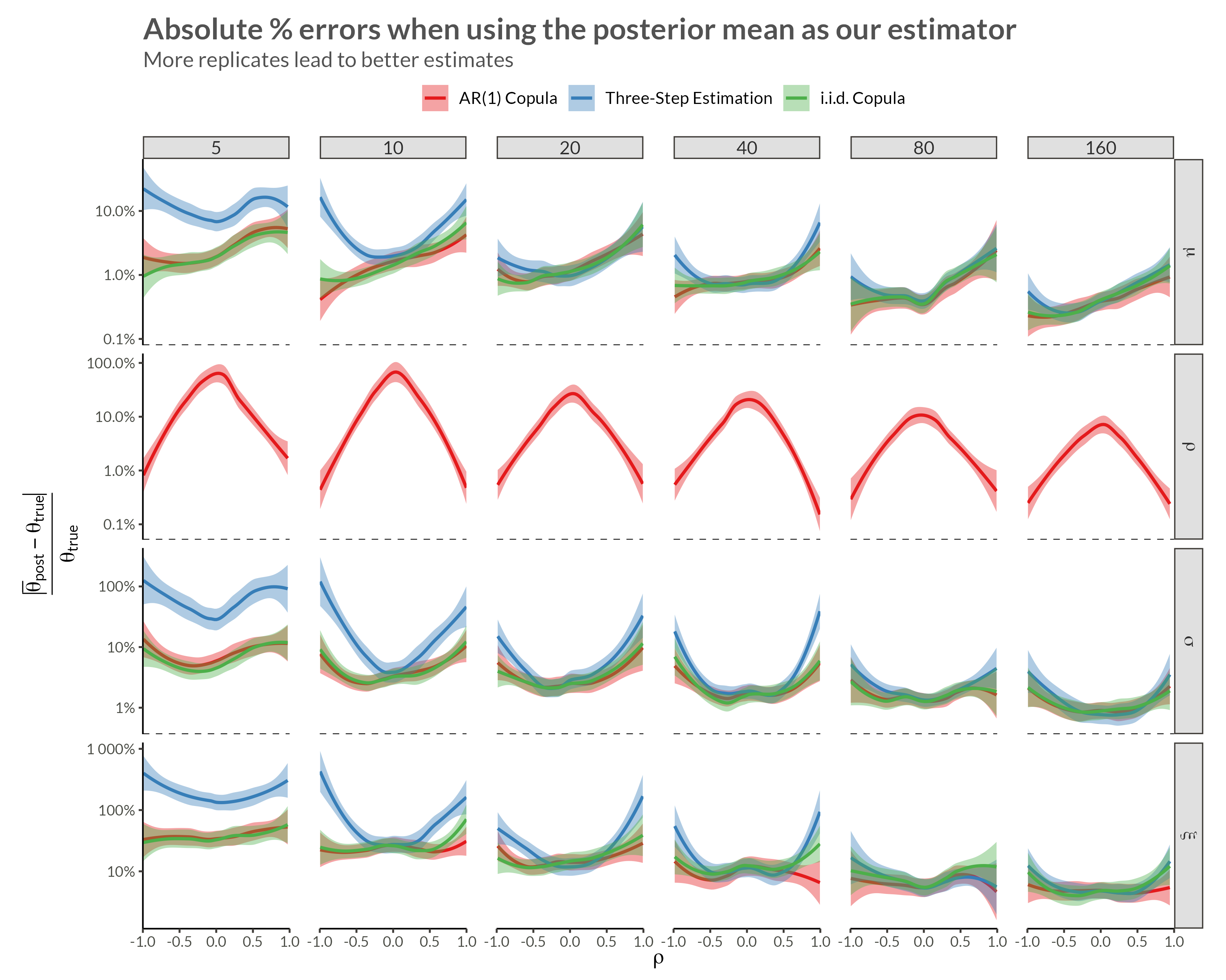
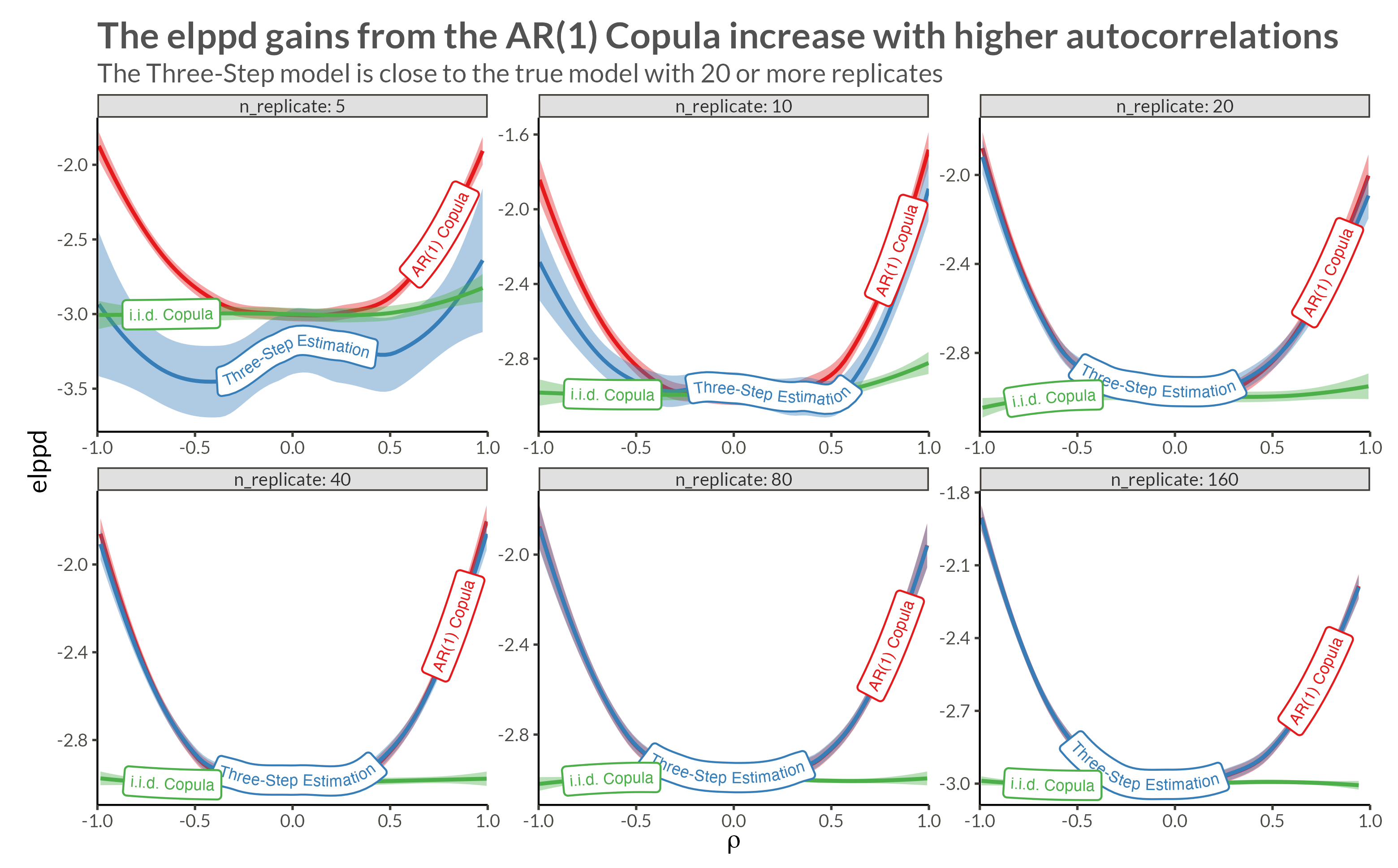
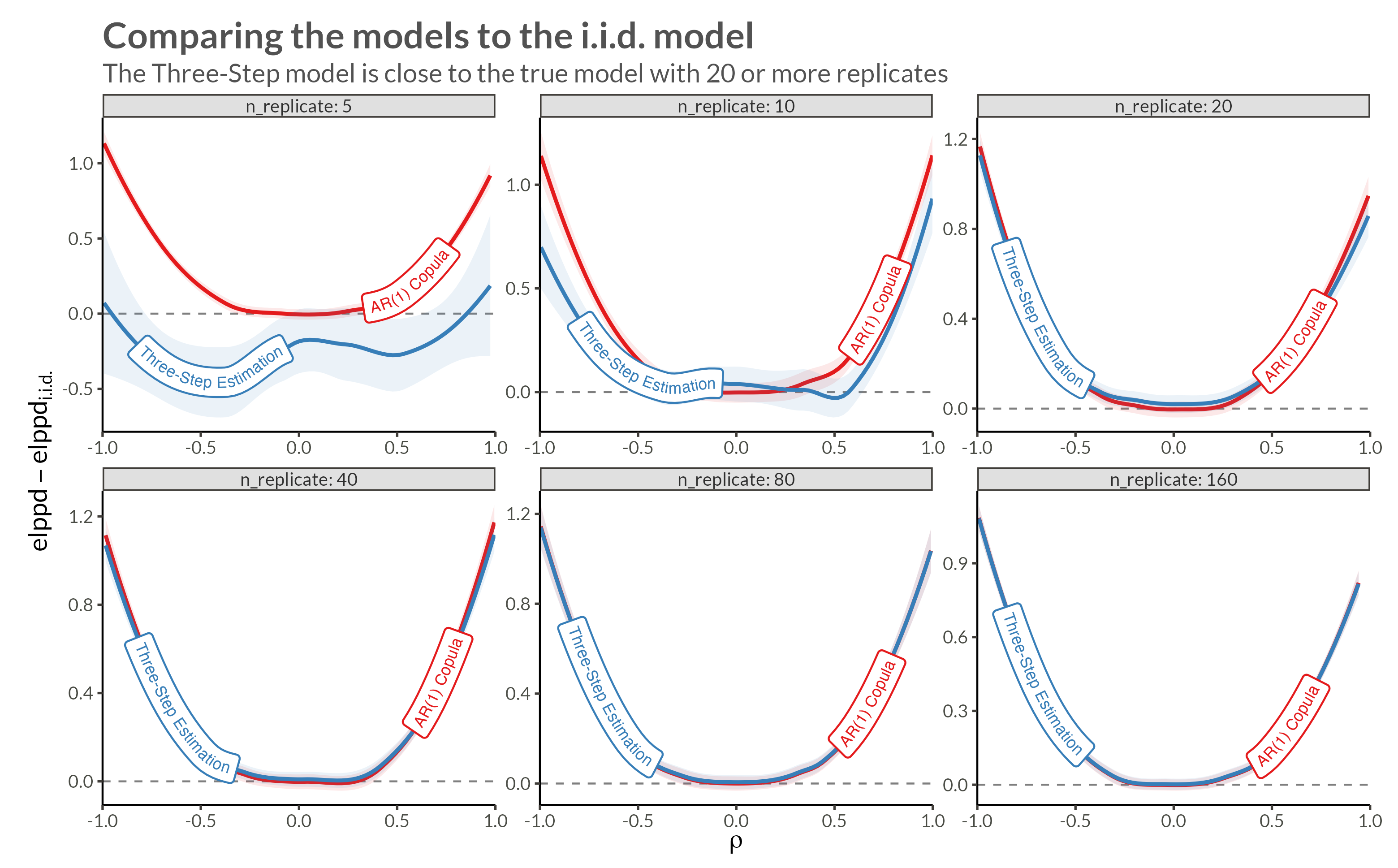
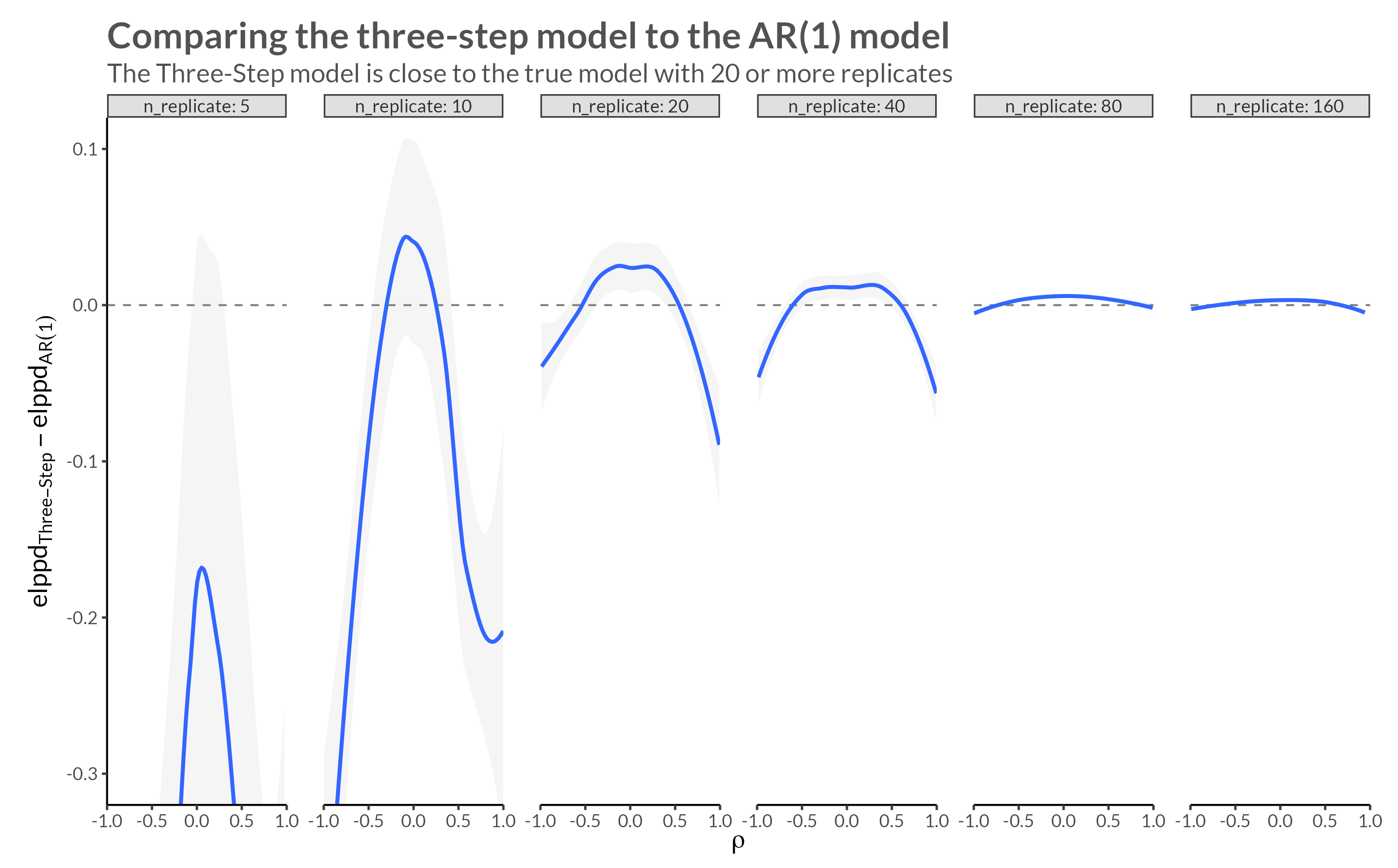
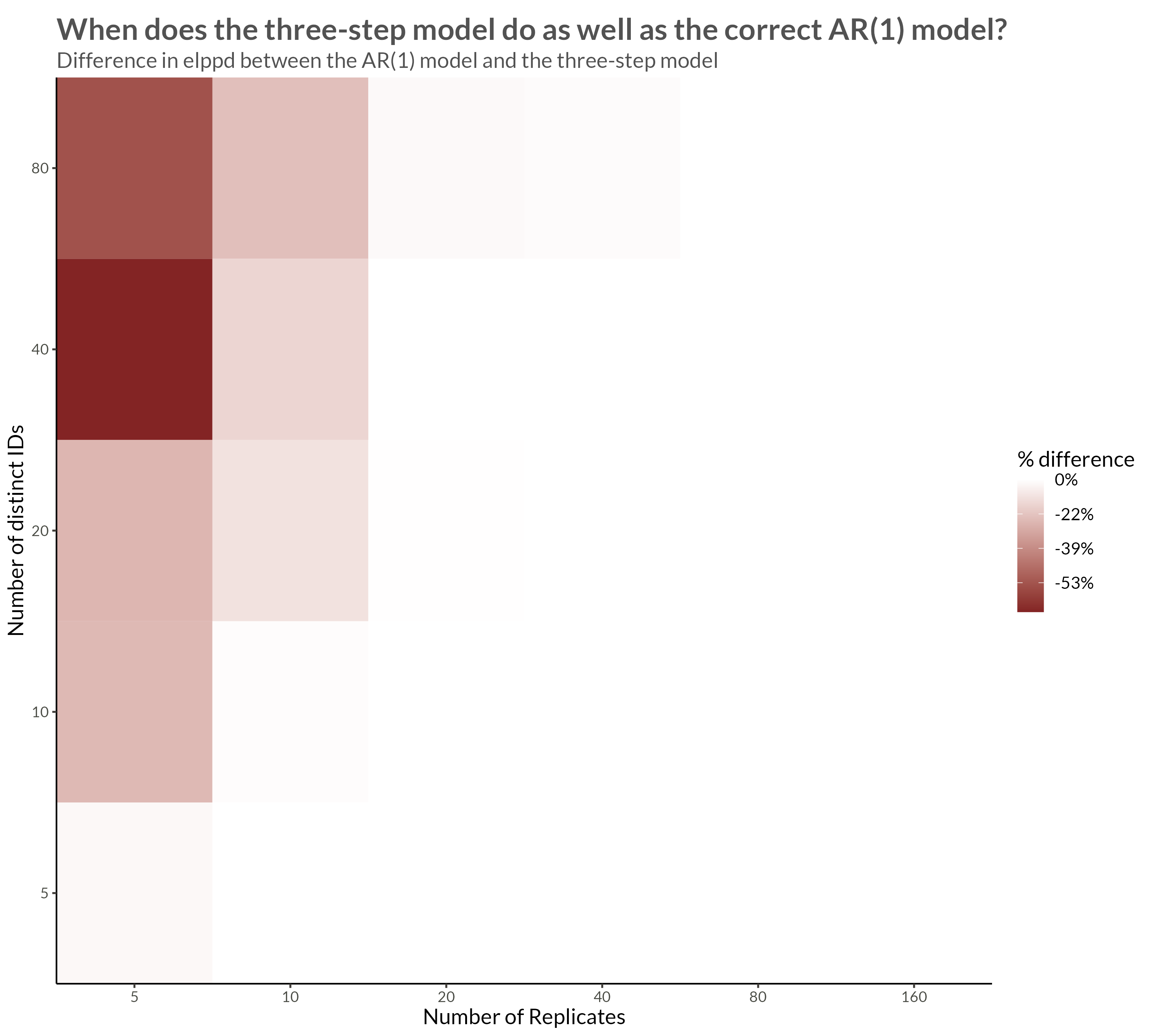
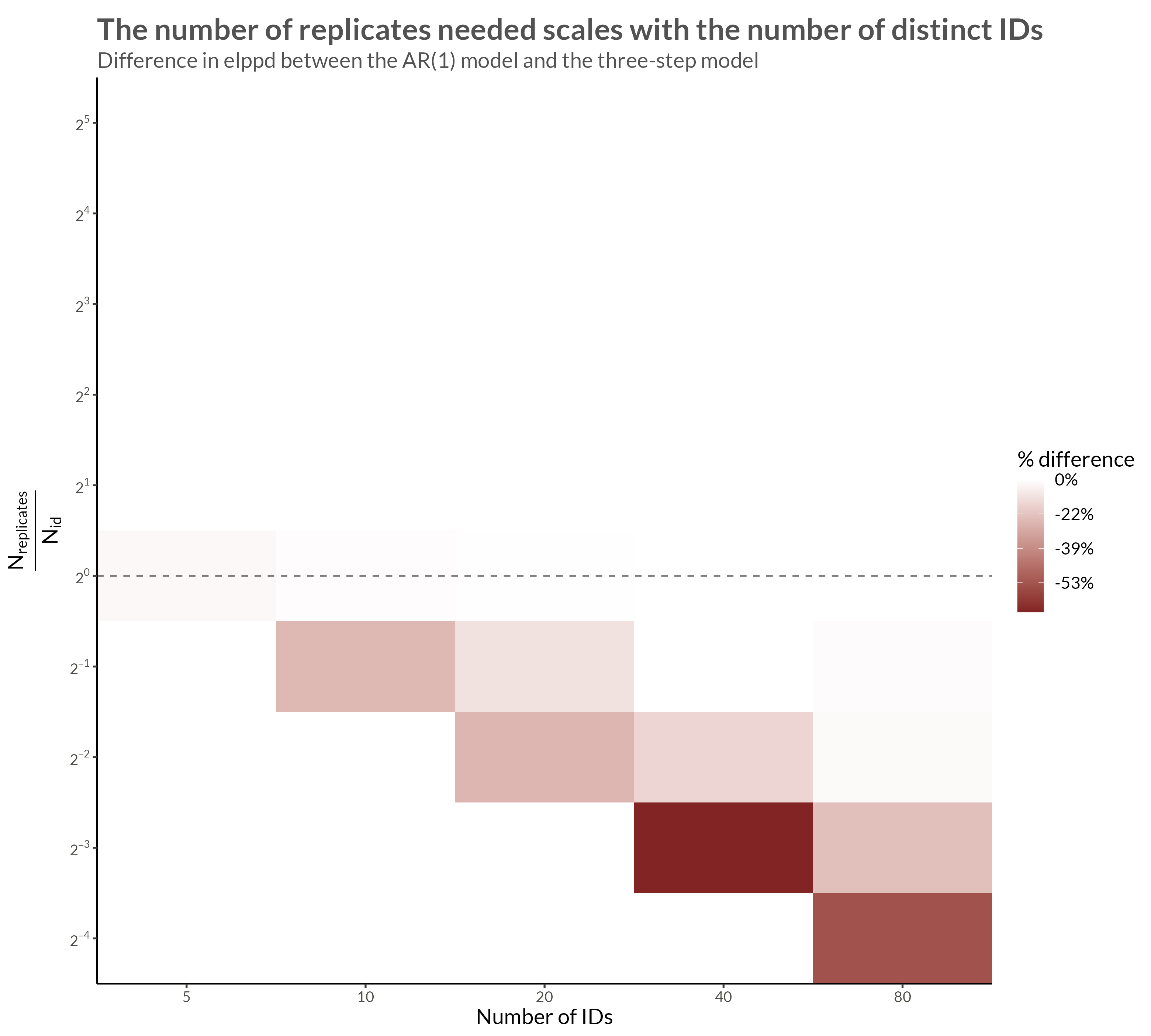
We see that with higher neighbor correlations we get obvious horizontal stripes corresponding to a large amount of dependence within each replicate.
We can fit the AR(1) model directly using Stan.
functions {
real normal_ar1_lpdf(vector x, real rho) {
int N = num_elements(x);
real out;
real log_det = - (N - 1) * (log(1 + rho) + log(1 - rho)) / 2;
vector[N] q;
real scl = sqrt(1 / (1 - rho^2));
q[1:(N - 1)] = scl * (x[1:(N - 1)] - rho * x[2:N]);
q[N] = x[N];
out = log_det - dot_self(q) / 2;
return out;
}
real normal_copula_ar1_lpdf(vector U, real rho) {
int N = rows(U);
vector[N] Z = inv_Phi(U);
return normal_ar1_lpdf(Z | rho) + dot_self(Z) / 2;
}
real gev_cdf(real y, real mu, real sigma, real xi) {
if (abs(xi) < 1e-10) {
real z = (y - mu) / sigma;
return exp(-exp(z));
} else {
real z = 1 + xi * (y - mu) / sigma;
if (z > 0) {
return exp(-pow(z, -1/xi));
} else {
reject("Found incompatible GEV parameter values");
}
}
}
real gev_lpdf(real y, real mu, real sigma, real xi) {
if (abs(xi) < 1e-10) {
real z = (y - mu) / sigma;
return -log(sigma) - z - exp(-z);
} else {
real z = 1 + xi * (y - mu) / sigma;
if (z > 0) {
return -log(sigma) - (1 + 1/xi) * log(z) - pow(z, -1/xi);
} else {
reject("Found incompatible GEV parameter values");
}
}
}
}
data {
int<lower = 0> n_replicate;
int<lower = 0> n_id;
array[n_replicate, n_id] real y;
array[n_replicate, n_id] real y_test;
}
parameters {
real<lower = 0> mu;
real<lower = 0> sigma;
real<lower = -0.5, upper = 1> xi;
real<lower = -1, upper = 1> rho;
}
model {
for (i in 1:n_replicate) {
vector[n_id] U;
for (j in 1:n_id) {
U[j] = gev_cdf(y[i, j] | mu, sigma, xi);
target += gev_lpdf(y[i, j] | mu, sigma, xi);
}
target += normal_copula_ar1_lpdf(U | rho);
}
}
generated quantities {
real log_lik = 0;
{
for (i in 1:n_replicate) {
vector[n_id] U;
for (j in 1:n_id) {
U[j] = gev_cdf(y_test[i, j] | mu, sigma, xi);
log_lik += gev_lpdf(y_test[i, j] | mu, sigma, xi);
}
log_lik += normal_copula_ar1_lpdf(U | rho);
}
}
}functions {
real gev_lpdf(real y, real mu, real sigma, real xi) {
if (abs(xi) < 1e-10) {
real z = (y - mu) / sigma;
return -log(sigma) - z - exp(-z);
} else {
real z = 1 + xi * (y - mu) / sigma;
if (z > 0) {
return -log(sigma) - (1 + 1/xi) * log(z) - pow(z, -1/xi);
} else {
reject("Found incompatible GEV parameter values");
}
}
}
}
data {
int<lower = 0> n_replicate;
int<lower = 0> n_id;
array[n_replicate, n_id] real y;
array[n_replicate, n_id] real y_test;
}
parameters {
real<lower = 0> mu;
real<lower = 0> sigma;
real<lower = -0.5, upper = 1> xi;
}
transformed parameters {
}
model {
for (i in 1:n_replicate) {
for (j in 1:n_id) {
target += gev_lpdf(y[i, j] | mu, sigma, xi);
}
}
}
generated quantities {
real log_lik = 0;
{
for (i in 1:n_replicate) {
for (j in 1:n_id) {
log_lik += gev_lpdf(y_test[i, j] | mu, sigma, xi);
}
}
}
}We can’t fit this model directly using one stan file. What we do is:
functions {
real gev_cdf(real y, real mu, real sigma, real xi) {
if (abs(xi) < 1e-10) {
real z = (y - mu) / sigma;
return exp(-exp(z));
} else {
real z = 1 + xi * (y - mu) / sigma;
if (z > 0) {
return exp(-pow(z, -1/xi));
} else {
reject("Found incompatible GEV parameter values");
}
}
}
real gev_lpdf(real y, real mu, real sigma, real xi) {
if (abs(xi) < 1e-10) {
real z = (y - mu) / sigma;
return -log(sigma) - z - exp(-z);
} else {
real z = 1 + xi * (y - mu) / sigma;
if (z > 0) {
return -log(sigma) - (1 + 1/xi) * log(z) - pow(z, -1/xi);
} else {
reject("Found incompatible GEV parameter values");
}
}
}
real normal_prec_chol_lpdf(vector x, array[] int n_values, array[] int index, vector values, real log_det) {
int N = num_elements(x);
int counter = 1;
vector[N] q = rep_vector(0, N);
for (i in 1:N) {
for (j in 1:n_values[i]) {
q[i] += values[counter] * x[index[counter]];
counter += 1;
}
}
return log_det - dot_self(q) / 2;
}
real normal_copula_prec_chol_lpdf(vector U, array[] int n_values, array[] int index, vector value, real log_det) {
int N = rows(U);
vector[N] Z = inv_Phi(U);
return normal_prec_chol_lpdf(Z | n_values, index, value, log_det) + dot_self(Z) / 2;
}
}
data {
int<lower = 0> n_replicate;
int<lower = 0> n_id;
array[n_replicate, n_id] real y;
array[n_replicate, n_id] real y_test;
int<lower = 0> n_nonzero_chol_Q;
real log_det_Q;
array[n_id] int n_values;
array[n_nonzero_chol_Q] int index;
vector[n_nonzero_chol_Q] value;
}
parameters {
real<lower = 0> mu;
real<lower = 0> sigma;
real<lower = -0.5, upper = 1> xi;
}
model {
for (i in 1:n_replicate) {
for (j in 1:n_id) {
target += gev_lpdf(y[i, j] | mu, sigma, xi);
}
}
}
generated quantities {
real log_lik = 0;
{
for (i in 1:n_replicate) {
vector[n_id] U;
for (j in 1:n_id) {
U[j] = gev_cdf(y_test[i, j] | mu, sigma, xi);
log_lik += gev_lpdf(y_test[i, j] | mu, sigma, xi);
}
log_lik += normal_copula_prec_chol_lpdf(U | n_values, index, value, log_det_Q);
}
}
}